scikit-learn Pipeline gotchas, k-fold cross-validation, hyperparameter tuning and improving my score on Kaggle's Forest Cover Type Competition
I spent the past few days exploring the topics from chapter 6 of Python Machine Learning, "Learning Best Practices for Model Evaluation and Hyperparameter Tuning". Instead of working through the exact same examples as the author I applied the learnings to another take at Kaggle's Forest Cover Type Dataset in a follow up notebook to my first attempt.
K-fold cross-validation
One of the first things you learn about in applying ML is the importance of cross-validation: evaluating the performance of your model on a portion of your dataset separate from what you used to train your model. The easiest way is to holdout a test set and compare performance using that:
- train your model on 70% of your labeled data
- evaluate the trained model on the remaining 30%
K-fold cross-validation improves on this by letting you do this multiple times so you can see whether the test performance varies based on which samples you used to train / test.
By running through the train/test comparison several times you can get a better estimate of model performance, and sanity check that the model is not performing wildly differently after being trained on different segments of your labeled data, which in itself could indicate instability in your model or too small a sample set.
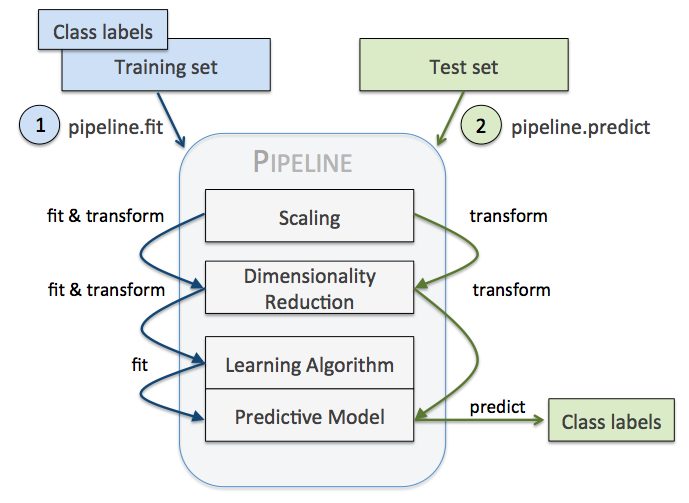
The need for Pipelines
As soon as you start doing k-fold cross validation it becomes very handy to make use of scikit-learn helper methods to do this grunt work for you. While hand rolling a 70/30 split isn't so bad, iterating over k different folds, tallying up the performances etc feels more like busy work.
But the question is, how can you package up something that represents all of the steps for use over and over again? This is where scikit-learn pipelines come in very handy.
The key point to me is that you can't just preprocess your entire labeled dataset once and then slice and dice it for cross-validation thereafter; preprocessing and dimensionality reduction make use of the training set too, so it is important that they only have access to whatever training set you are using during the k-fold process, otherwise you are granting some of your data processing steps access to "unseen" data and aren't really testing how it will fair in the wild with new unseen data.
So if I need to pass along something that will fit / transform / predict, I need to construct a pipeline, which is a composite that implements the same interface as all of the learning models:
- fit: given samples, tune some internal parameters
- transform: return some transformation of samples, possibly making use of anything you tuned while fitting
In addition to any number of fit/transform steps, pipeline's implement a predict method that use the last item in the pipeline, which it expects to be a model.
So in order to participate in this I needed refactor my hand rolled preprocessing function into one that constructs a pipeline.
This post was very handy in finding the FeatureUnion pipeline that can help you break down your preprocessing and recombine it again. So you can combine two pipelines:
- one that extracts categorical variables and applies one hot encoding, and then scales the outputs to -1 / 1
- one that extracts quantitative variables, imputes missing values with each variable's mean and then applies mean scaling into one and have a little pipeline that is ready to preprocess your data. This can then be further combined in a larger pipeline with other steps like PCA followed by your classifier.
Pipeline gotchas
Pipeline's are very cool but can still be a little clunky to work with. I found that when parallelization happens behind the scenes, any hand-rolled Pipeline step needs to extend BaseEstimator to inherit some magic parameter serialization logic and that you need to be careful to have anything that needs to persist between fit and transform to be declared a parameter in the _init_ method of the class in order to work. This isn't really documented anywhere outside of blog posts and stack overflow questions.
Hyperparameter tuning
Having each model I wanted to test all rolled up into tidy pipelines, I was ready to make use of some other cool stuff in scikit-learn, including hyperparemter tuning with grid search, which will evaluate the performance of a model with varying parameter values using k-fold cross validation along the way. Note: "hyperparameter" is just a fancy name for the parameters used to tune the models themselves. For instance, the maximum depth you allow a decision tree to grow to is a hyperparameter.
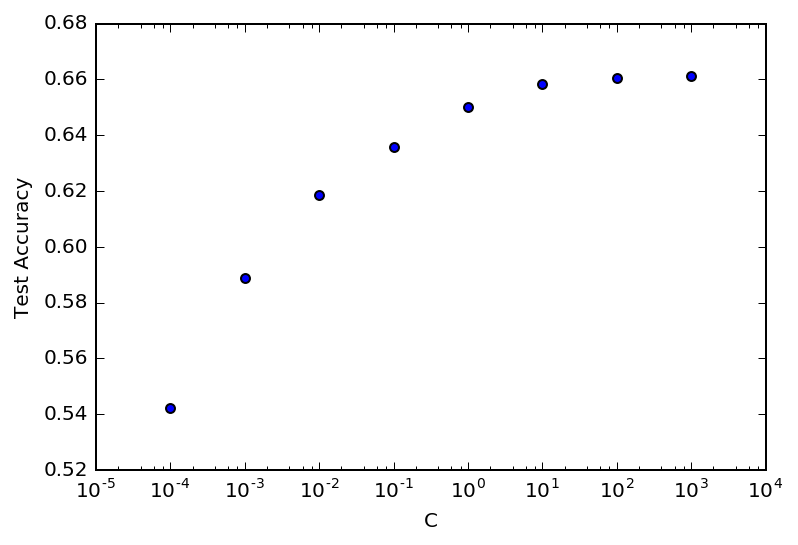
Here is a graph of how model performance varied across the tested parameters for Logistic regression:
Note that C is the inverse regularization parameter, so increasing it reduces the dampening of parameter values, allowing the weights of the trained model vary as much as they like to fit the data. In this case, LR was not going to overfit and it was best to essentially remove all regularization.
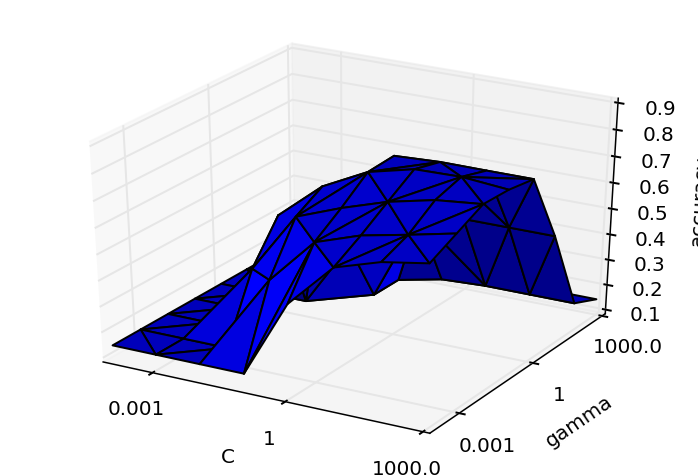
Kernel SVM:
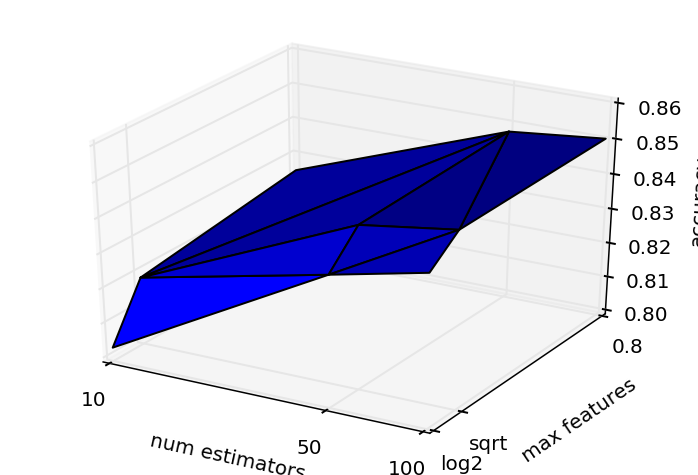
and Random Forest:
for once, these graphs were produced via original work, not copy/paste from Python ML examples :) What strikes me is that: yes these parameters matter, and also that in these cases, the performance across parameter space is convex, making me wonder whether it could be worth applying something smarter than exhaustive search? As this post notes most people don't bother as it makes it harder to parallelize the search across parameters, and instead recommends randomized search.
Conceptually, hyperparameter tuning is an optimization task, just like model training... Smarter tuning methods are available. Unlike the “dumb” alternatives of grid search and random search, smart hyperparameter tuning are much less parallelizable. Instead of generating all the candidate points upfront and evaluating the batch in parallel, smart tuning techniques pick a few hyperparameter settings, evaluate their quality, then decide where to sample next. This is an inherently iterative and sequential process. It is not very parallelizable. The goal is to make fewer evaluations overall and save on the overall computation time. If wall clock time is your goal, and you can afford multiple machines, then I suggest sticking to random search.
Parallelization gotchas
In evaluating many possible parameters across k folds, you can easily ramp up to 60+ train/fit runs on a given model, which can be really slow, especially on SVM models. Scikit-learn let's you run all of this across multiple cores to speed things up via the njobs param, and this worked well until I ran it during the hyperparameter tuning of random forest classifiers; running multiple 100 tree models in parallel crashed the scikit-learn cell and I had to just kind of guess that this was what was happening and finding that for that case, setting njobs=1 worked, I just needed to let it run for about 15 minutes to complete and find the optimal parameters.
Improving performance on Kaggle
After all of this futzing around, how did the improved parameters fair? The adjustments were enough to get me a 1-3% boost.
While this is kind of underwhelming, it was nice to see something concrete and this process can always help you if you happen to have guessed a really bad parameter value: the graphs above show that there were much worse values I could have chosen in the LR and SVM models.
| Model | Untuned Test Accuracy | Tuned Test Accuracy | Untuned Kaggle Score | Kaggle Score |
|---|---|---|---|---|
| Logistic Regression | 0.658 | 0.658 | 0.56 | |
| Kernel SVM | 0.815 | 0.825 | 0.72143 | 0.73570 |
| Random Forest | 0.82 | 0.85 | 0.71758 | 0.74463 |